Predominance of metallo-beta-lactamase blaVIM Genes in Clinical Isolates from Health Facilities in Nigeria: Role in Multidrug Resistance Surge
1Ubong Ekerenam Etang, 2Anietie Effiong Moses, 3Samuel Sunday Akpan, 4Aniekan-Augusta Okon Eyo, 5Ekemini Anietie Moses
Abstract
Background: The predominance of metallo-beta-lactamase (MBL) genes in Gram-negative bacteria (GNB) has been reported to contribute immensely in the emergence of multi-drug resistance (MDR) strains in hospital environments worldwide.
Aim: This study aimed to investigate the molecular characteristics of MBL genes in MDR-GNB in selected health facilities in Akwa Ibom State, Nigeria.
Materials and methods: A descriptive cross-sectional study was conducted in three health facilities in the State. One hundred and sixty (160) samples each of wound, urine and blood were collected aseptically from consented patients and analyzed using standard laboratory procedures. Identification, antibiotic susceptibility testing and phenotypic detection of MBL-producers were done using the VITEK®2 system, Kirby-Bauer disc diffusion and imipenem+EDTA CDT, respectively. Polymerase chain reaction was used to investigate the characteristics of MBL encoding genes: blaVIM, blaNDM and blaIMP.
Results: Out of 135 GNB identified, 60.7% exhibited MDR and 53(39.3%) were MBL-producers. Of the MBL genes screened, only the blaVIM was detected in 5(23.8%) of the 21 selected strains screened. The breakdown of blaVIM gene detection rate among isolates from wound, blood and urine samples were 20%, 11.1% and 3.4%, respectively. The GNB that harboured the blaVIM genes were Serratia marcescens, Klebsiella pneumoniae and Escherichia coli recovered from wounds as well as Proteus mirabilis and Pseudomonas aeruginosa recovered from blood and urine samples, respectively.
Conclusion: Findings of this study highlight the importance of the blaVIM gene in conferring multidrug among the Gram-negative bacteria, and the need to establish antimicrobial resistance surveillance network and policy to determine appropriate empirical treatment regimen among hospitals in the State.
Keywords: Metallo-beta-lactamase, Gram-negative bacteria, Multi-drug resistance, blaVIM genes
Introduction
The emergence of multidrug resistance (MDR) in clinically important Gram-negative pathogens poses a serious public health threat in the treatment of related infections globally.1 Antibiotics that belong to the beta-lactam class have been used successfully over the last 6 decades, and genes that code for the production of metallo-beta-lactamase (MBL) enzymes have emerged in some strains of bacteria.2,3 These resistance genes which are disseminated within a bacterial population through horizontal gene transfer (HGT) mechanisms have been implicated in delayed therapeutic outcomes, allowing infections to progress despite antibiotic administration.4,5
Metallo-beta-lactamases (MBLs) are class B group of beta-lactamases that catalyze the hydrolysis of a broad range of beta-lactam drugs (such as penicillins and their derivatives, cephalosporins including those with oxyimino side chain, cephamycins and oxapenems), and the cabapenems.6,7 The MBL enzyme types, grouped on the basis of amino acid sequence homology include the Verona integron-encoded metallo-beta-lactamases (VIM), Imipenemase (IMP), Sau Paulo metallo-beta-lactamase (SPM), German Imipenemase (GIM), New Delhi metallo-beta-lactamase (NDM), Seoul Imipenemase (SIM) and Dutch Imipenemase (DIM).8,9 The catalytic ability of these enzymes, which require zinc-ions, is not neutralized by commercially available beta-lactamase inhibitors such as clavulanate, tazobactam and sulbactam.10 The genes that code for MBLs are carried on plasmids, transposons and gene cassettes which are the mobile genetic elements. This acquisition is responsible for the evolution and spread of multi-drug resistant (MDR) bacterial phenotypes through vertical or horizontal gene transfer mechanism.11
Among the MBLs, carbapenemases are of significant concern due to their increased efficacy against carbapenems, the broad-spectrum drugs often used for treating life-threatening infections, or as the empirical agents of last resort for treating infections caused by multi-drug resistant bacteria. This further limits the therapeutic option available in the management of infections caused by MDR Gram-negative bacteria pathogens.2
Currently, clinically active MBL inhibitors are readily not available and the easy spread of genes that encode them remains the most significant cause of multiple resistances seen in Gram-negative bacteria (GNB).10 In view of the fact that, these MBL-borne pathogens are implicated in various hospital and community-acquired infections, application of molecular procedures to detect and characterize the different MBL genes and their MDR profiles becomes indispensable in providing useful epidemiological data that are necessary for the implementation of effective control policy and management of infections.5
Studies have reported a high incidence of metallo-beta-lactamases and their involvement in the widespread dissemination of multi-drug resistance globally, especially among Gram-negative clinical isolates.5,12 The prevalent MBL genes vary in different countries and regions of the world. For instance, MBLs with blaVIM and blaNDM have been reported in Khartoum hospitals in Sudan as the prevalent MBL genes;13 blaIMP in Bayelsa State, Nigeria14 and in Nadu, India;15 blaVIM in Abuja, Nigeria7 as well as blaVIM and blaIMP in Italy16 and Malaysia.17 However, in Akwa Ibom State, Nigeria, information regarding the cause of multi-drug resistance surge in hospitals, the dominant MBL gene types in circulation as well as their transmission dynamics in clinical isolates still remains limited. Therefore, this study was carried out to investigate the molecular characteristics of the metallo-beta-lactamase genes in multi-drug resistant Gram-negative isolates from selected health facilities in Akwa Ibom State, Nigeria.
Materials and Methods
Study design and population
This was a descriptive cross-sectional study carried out in three health facilities namely: University of Uyo Teaching Hospital (UUTH), Uyo; General Hospital, Ikot Ekpene (GHIE) and Immanuel Hospital, Eket (IHE), located in 3 different senatorial districts in Akwa Ibom State from November 2021 to June 2023. The study population comprised 480 patients who had been previously diagnosed or suspected of bacteremia, wound infection and urinary tract infections. Clinical samples were collected from the subjects after obtaining informed consent.
Ethical considerations
Approval to conduct the study was obtained from Health Research Ethics Committees (HRECs) of the Akwa Ibom State Ministry of Health (AKSMH), with HREC assigned No: AKHREC/27/8/21/008 and University of Uyo Teaching Hospital (UUTH), Uyo with HREC assigned No: UUTH/AD/S/96/VOL.XXI/629. Written informed consent was obtained from participants prior to their inclusion in the study.
Identification of bacterial isolates
Gram-negative bacteria were recovered from wound, urine and blood samples of patients in the selected hospitals. Bacterial cultures were grown in MacConkey agar, differentiated by Gram staining technique, identified using the conventional biochemical methods and validated by VITEK®2 automated biochemical system according to standard protocols.
Antibiotic susceptibility testing (AST)
The antibiotic susceptibility testing of the isolates was carried out using Kirby-Bauer disc diffusion method after overnight incubation at 37oC on Mueller-Hinton agar plates as recommended by the Clinical Laboratory Standard Institute.18 The antibiotics with their concentrations included gentamicin (10µg), imipenem (10µg), amoxicillin-clavulanic acid (amoxicillin 20µg/clavulanate 10µg), ciprofloxacin (10µg), cefotaxime (30µg), ceftazidime (30µg), aztreonam (30µg), cefepime (30µg), trimethoprim-sulfamethoxazole (2.5µg), ceftriaxone (30µg) and ofloxacin (5µg) (Oxoid, UK). Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603 and Pseudomonas aeruginosa ATCC 27853 were used as reference strains for susceptibility testing, while AST results were interpreted in accordance with the CLSI guidelines and interpretive criteria.18
Phenotypic screening for MBL production
Gram-negative isolates that were resistant to imipenem at the initial screening test were selected for phenotypic confirmation of MBL production using the imipenem+EDTA combined disc test (IMP+EDTA CDT) according to previously described method.19 Overnight culture of the test isolates adjusted to 0.5 McFarland turbidity standard and inoculated on a Mueller-Hinton agar plate was used to screen for MBL production. Two imipenem discs (10µg), of which one was supplemented with 10µl of 0.5M EDTA solution were placed at a distance of 5cm apart and the plates incubated at 35oC for 16-18 hrs. Isolates with enhanced zone size of =7mm towards IMP+EDTA disc compared to IMP disc alone was considered and interpreted as MBL producers.19
Bacterial genomic DNA extraction and quantification
Extraction of bacterial DNA was done using a ZR fungal/bacterial DNA mini prep extraction kit supplied by Inqaba South Africa according to the manufacturer’s instruction. The extracted genomic DNA was quantified using the Nanodrop 1000 spectrophotometer (Thermo Scientific, Inqaba Biotec).
Amplification of IMP gene
The blaIMP gene from the isolates was amplified using the forward (5’–GAAGGCGTTTATGTTCATAC–3’) and reverse (5’ – GTATGTTTCAAGAGTGATGC – 3’) primers on a nexus gradient Mastercycler (Eppendorf, Germany) at a final volume of 30 µL for 35 cycles. The PCR mixture included: The X2 Dream Taq Master Mix supplied by Inqaba, South Africa (Taq polymerase, DNTPs, buffer, MgCl2), the primers at a concentration of 0.5 µM, 2 µL of the extracted DNA as template and 11.8 µL of PCR water. The PCR conditions were as follows: Initial denaturation, 95oC for 5 min, denaturation, 95oC for 30 sec; annealing, touchdown of 55-49oC for 30 sec; extension, 72oC for 30 sec for 35 cycles, final extension, 72oC for 5. The amplicon was resolved on a 1% agarose gel at 200V for 15 min and visualized on a blue light trans-illuminator but there was no base pair of product size.
Amplification of NDM gene
The blaNDM gene from the isolates was amplified using the forward (5’–GGTTTGGCGATCTGGTTTTC – 3’) and reverse (5’ –CGGAATGGCTCATCACGATC – 3’) primers on a nexus gradient Mastercycler (Eppendorf, Germany) at a final volume of 30 µL for 35 cycles. The PCR mixture included: The X2 Dream Taq Master Mix supplied by Inqaba, South Africa (Taq polymerase, DNTPs, buffer, MgCl2), the primers at a concentration of 0.5 µM, 2 µL of the extracted DNA as a template and 11.8 µL of PCR water. The PCR conditions were as follows: Initial denaturation, 95oC for 5 min, denaturation, 95oC for 40 sec, annealing, 56oC for 40 sec, extension, 68oC for 40 sec for 35 cycles, final extension, 68oC for 5 min and hold at 10min. The amplicon was resolved on a 1% agarose gel at 200V for 15 min and visualized on a blue light trans-illuminator but there was no base pair of product size.
Amplification of VIM gene
The blaVIM gene from the isolates was amplified using the forward (5’ – GTTTGGTCGCATATCGCAAC – 3’) and reverse (5’ – CTACTCGGCGACTGAGCGAT – 3’) primers on a nexus gradient Mastercycler (Eppendorf, Germany) at a final volume of 30 µL for 35 cycles. The PCR mixture included: The X2 Dream Taq Master Mix supplied by Inqaba, South Africa (Taq polymerase, DNTPs, buffer, MgCl2), the primers at a concentration of 0.5 µM, 2 µL of the extracted DNA as a template and 11.8 µL of PCR water. The PCR conditions were as follows: Initial denaturation, 95oC for 5 min, denaturation, 95oC for 30 sec, annealing, 57oC for 40 sec, extension, 72oC for 40 sec for 35 cycles, final extension, 72oC for 5 min and hold at 10 min. The amplicon was resolved on a 1% agarose gel at 200V for 15 min and visualized on a blue light trans-illuminator for a 265 base pair product size.
Statistical analysis
The statistical analysis was performed using SPSS software version 25.0 (IBM Corp., USA). Results obtained were presented as descriptive statistics in tables and percentages.
Results
The comparative in vitro antibiotic resistance patterns of MBL and non-MBL-producing Gram-negative clinical isolates in Akwa Ibom State is presented in Table 1. The results showed the highest level of resistance of MBL-producers against trimethoprim-sulfamethoxazole and ceftriaxone (81.1%), each. For the non MBL-producing isolates, in vitro resistance was observed against trimethoprim-sulfamethoxazole (76.8%), ceftriaxone (69.5%) and aztreonam (69.5%) antibiotics.
Table 1: In vitro antibiotic resistance patterns of MBL and non-MBL-producing clinical isolates in three health facilities in Akwa Ibom State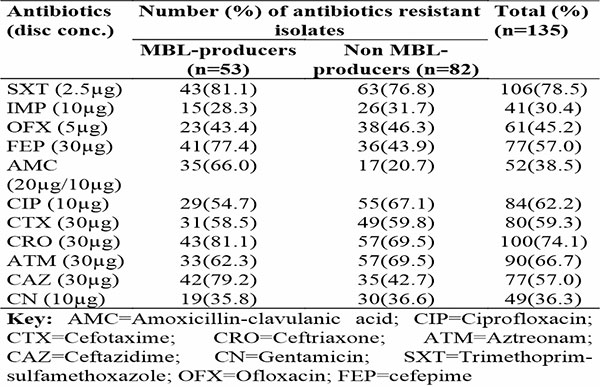
The percentage distribution of multidrug resistance (MDR) among MBL and non-MBL producing Gram-negative bacteria is presented in Figure 1. The results showed that all MBL-producing isolates (100%) were multidrug resistant while 35.4% of non-MBL producers were multidrug resistant. The overall prevalence of MDR Gram-negative bacteria in the three hospitals in Akwa Ibom State was 60.7%. An isolate was considered MDR if it resisted at least 3 different classes of antibiotics tested.
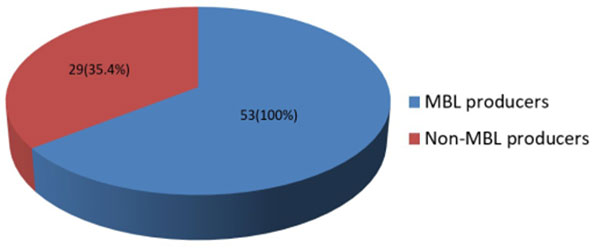
Figure 1: Percentage distribution of multidrug resistance (MDR) among MBL and non-MBL producers
The distribution of MBL genes in Gram-negative bacteria isolated from the patients is indicated in Table 2. Out of the three MBL genes assayed in 21 clinical isolates of Gram-negative bacteria, only blaVIM was detected in 5 of the isolates. The following GNB harboured the blaVIM genes: Serratia marcescens, Klebsiella pneumoniae, Proteus mirabilis, Escherichia coli and Pseudomonas aeruginosa.
Table 2: Distribution of MBL genes in Gram-negative bacteria isolated from the patients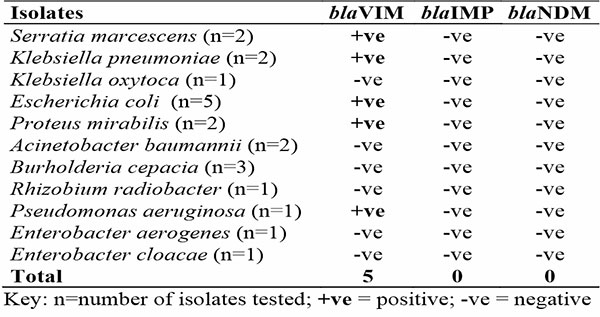
The distribution of blaVIM MBL positive genes in clinical samples in Akwa Ibom State is shown in Table 3. Of the 160 samples each of urine, blood and wound screened for the detection of MBL genes, the presence of blaVIM gene was in the rate of 3.4%, 11.1% and 20%, respectively. Hence, the overall prevalence of blaVIM gene detection in Akwa Ibom State was 23.8%.
Table 3: Distribution of blaVIM MBL positive genes according to clinical samples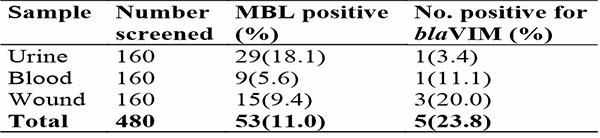
The agarose gel micrograph of blaVIM gene in MBL-producing Gram-negative isolates is shown in Figure 2. Lanes 4, 8, 11, 13, and 17 revealed the 265 base pairs of amplified blaVIM genes in S. marcescens, P. mirabilis, K. pneumoniae, E. coli and P. aeruginosa isolates, respectively. Lane U represents the 100bp DNA ladder.
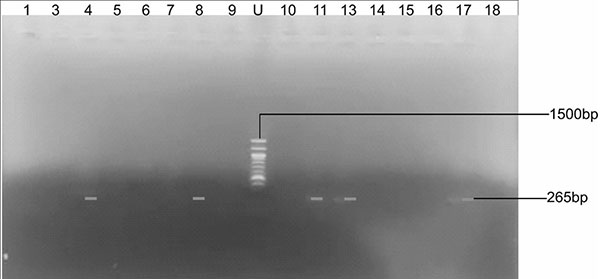
Figure 2: Agarose gel micrograph of blaVIM gene from MBL-producing isolates
Discussion
The preponderance of multi-drug resistance (MDR) Gram-negative bacteria isolates in clinical samples is worrisome, especially in the area of therapeutic management of infections caused by these etiologic agents.1,5 The predominance of blaVIM genes, among other MBL gene types in different parts of the world have been reported to contribute immensely in the widespread emergence of MDR strains in hospital environments.12,20 The situation is even compounded by the absence of molecular diagnostic facilities for accurate detection of these resistance genes, particularly in low-resource settings, including Nigeria. In this study, 15(28.3%) out of the 41 imipenem-resistant Gram-negative bacteria produced MBLs. Among the folate pathway antagonist group, resistance was highly detected in MBL-producing isolates against trimethoprim-sulfamethoxazole (SXT: 81.1%), while for the cephalosporin group, ceftriaxone (CRO: 81.1%) and ceftazidime (CAZ: 79.2%) showed high resistance rates. However, for the carbapenem group, a low resistance rate of 28.3% was obtained among MBL-positive isolates. These findings corroborate earlier reports from Emam Hossein and Loghman Hakim hospitals in Japan,21 Immanuel Hospital, University of Uyo Teaching Hospital and General Hospital Ikot Ekpene in Akwa Ibom State, Nigeria5 and a tertiary health facility in South India.22 The production of these enzymes in certain Gram-negative clinical isolates has been reported in many studies to have contributed to the increased development of acquired resistance to both beta-lactam and non-beta-lactam antibiotics.1,23,24
Multidrug resistance (MDR) is a bane in many hospitals as it poses a significant therapeutic challenge in the management of infections.1,25 This study revealed 60.7% overall prevalence of multidrug resistance Gram-negative bacteria in the three health facilities. All MBL-producing Gram-negative clinical isolates (100%) were multidrug resistance portraying the significant impact of MBL in the evolution of multidrug resistant strains. The prevalence of MDR obtained in this study is higher than that reported in previous studies.25,26 The possible reason for this high prevalence may not be unconnected to irrational use of antibiotics, over-the-counter prescription by non clinicians and absence of antibiotic sensitivity testing of clinical isolates prior to antibiotics prescription by physicians.5
In this study, of the 21 selected MBL-producing Gram-negative isolates, 5 (23.8%) were found to harbor blaVIM gene while the other MBL genes, blaIMP and blaNDM were not detected in any of the clinical isolates tested. This contradicts the findings of Akereuke et al.27 in which all the above mentioned 3 genes were detected among Gram-negative isolates recovered from inpatients with urinary tract infection (UTI) and wound infection in University of Uyo Teaching Hospital, being one of the health facilities used in this study. The reason for this observed difference is not clear, but could be due to the primers used in studies. This report is also inconsistent with the findings of Abdu et al.28 in which both NDM and VIM genes were detected among clinical isolates of P. aeruginosa and K. pneumoniae, while only IMP gene was not detected in Bayelsa State, Nigeria. However, it is consistent with the findings of Zubair et al.7 in Abuja, in which only blaVIM gene was detected out of blaIMP, blaNDM, blaSPM, blaGIM and blaSIM among Pseudomonas aeruginosa isolates. A recent study in Bayelsa State, Nigeria by Alade et al.14 did not detect NDM and VIM genes in clinical isolates of P. aeruginosa isolates and Davasena et al.15 did not detect NDM-1 gene in P. mirabilis and P. aeruginosa isolates in a study conducted in Tamil, Nadu, India. Those bacterial isolate that resisted carbapenems but negative for any of the three MBL genes tested may have acquired other MBL genes which were not assayed in this study. Resistance may have been due to other mechanisms of antibiotic resistance such as outer membranes’ impermeability, target sites’ modification, efflux pump and possession of other carbapenemases.14
Also, it has been documented in literature that detection of MBL genes greatly vary among regions and countries of the world, due to differences in circulating strains, genetic variability and environmental factors.29,30 Organism-specific gene variation may as well vary within country as observed in this study. MBL genes previously identified in a recent study among E. coli isolates in Anambra State University Teaching Hospital, Awka, Southeastern Nigeria include VIM, IMP, SPM, SIM and GIM-gene types.31 The carbapenem resistant gene, blaIMP was detected among P. aeruginosa clinical isolates in Benin City, Nigeria,32 NDM genes were detected in some E. coli isolates in Nnamdi Azikiwe University Teaching Hospital (NAUTH), Awka, Southeastern Nigeria and NDM-1 gene was responsible for the resistance of some E. coli and K. pneumoniae isolates to carbapenem in Kano, Northern Nigeria.33 Higher detection rates for MBL genes for some organisms have also been reported in studies outside Nigeria. For instance, blaSIM (33%) in E. coli isolates from Indian hospital;34 IMP-1, IMP-7, IMP-11 and VIM-1 genes detected among P. aeruginosa isolates from Japanese hospital;35 blaIMP (36.8%) and blaVIM (15.7%) genes among E. coli isolates from hospitals in Malaysia;17 VIM (38.9%), IMP (26.4%) and NDM (4.2%) genes among clinical isolates of Citrobacter spp, Escherichia coli, Enterobacter spp, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Proteus vulgaris from Khartoum hospitals in Sudan between 2015 and 2016;13 blaIMP-1 (3.48%) and blaVIM-1 (17.44%) genes among Acinetobacter baumannii isolates from two hospitals in Tehran, Iran;36 and VIM and IMP-type enzymes among MBL-producing Pseudomonas aeruginosa, Pseudomonas putida and Enterobacter cloacae isolates from a countrywide survey in Italy.16
According to Lauretti et al., the VIM, IMP and NDM-type genes are the most studied MBLs, first detected in the 1990s. The VIM gene was first isolated from P. aeruginosa strain in Verona Hospital, Italy in 1997.37 It has now been recovered from many infections such as urinary tract infections, wound infections, blood-borne infections and device-associated infections.7,34 Currently, these genes have been detected in more than 23 Gram negative bacterial species including P. aeruginosa, Acinetobacter spp. and members of Enterobacteriaceae family across 40 countries/regions of the world, largely in Europe, Asia, America and the Mediterranean region.6,7 The predominance of the VIM gene in isolates obtained from this study may not be unconnected with cases of imported antibiotic resistance from Israel, Rome and Saudi Arabia by Nigerian travelers for pilgrimage and international travel for medical tourism and work as reported by Zubair et al.7 There are presently 46 VIM variants (VIM-1 to -46) documented. While VIM-1 has 31.4% amino acid sequence identity with IMP-1, VIM-2 is the most widely distributed MBLs that have been implicated in multiple outbreaks of MBL-mediated antibiotic resistance globally.38
In this study, the blaVIM genes were detected in three Gram-negative bacterial strains recovered from wound infections (Serratia marcescens, Klebsiella pneumoniae and Escherichia coli) and one each from blood stream infection caused by Proteus mirabilis and UTI caused by Pseudomonas aeruginosa. Similarly, Abdu et al.28 reportedly detected 6 isolates of Gram-negative bacteria (GNB) habouring the VIM genes from patients with various infections in Bayelsa. Four (4) P. aeruginosa isolates were recovered from wound infection (66.7%) while one each of E. coli and K. pneumoniae were from UTI (33.3%). To the best of our knowledge, rarely is carbapenem-resistant Serratia marcescens reported in Nigeria and this is the first report on blaVIM gene detection in this bacterium in Akwa Ibom State. Most published studies on beta-lactamases detection among Gram negative clinical isolates in Akwa Ibom State were limited to ESBLs.39,40,41 Only one study in UUTH reported on molecular detection of MBL genes in Gram-negative clinical isolates, in which the blaVIM gene was majorly detected in P. mirabilis, K. pneumoniae, E. coli and P. aeruginosa isolates.27 This suggests that the blaVIM genes are probably the dominant MBL-gene type circulating in clinical isolates of Gram-negative bacteria in Akwa Ibom State.
Conclusion
The blaVIM MBL-positive Gram-negative bacteria are predominantly in circulation among bacteria causing multidrug resistant infections in patients seen in hospitals in Akwa Ibom State. This study is the first to report the detection of blaVIM gene in clinical isolates of carbapenem-resistant Serratia marcescens in wound samples. The high prevalence of blaVIM gene detection in this study (23.8%) is perturbing and calls for the establishment of systematic surveillance network and initiation of policies for monitoring of patients with multi-drug resistant Gram-negative bacteria infections (MDR-GNBIs) in the State. This is pertinent because the spread of these resistant genes in clinical isolates if left unchecked, could lead to widespread dissemination of uncontrollable epidemic clones for which efficient antimicrobial regimen may not be readily available.
Acknowledgments
The authors are thankful to Prof. Tatfeng Mirabeau and his staff for their assistance during molecular laboratory investigation. We also acknowledge the technical assistance of Susan Adie in the course of the biochemical identification of GNB with VITEK®2 system.
Conflict of Interest
The authors declared that there was no conflict of interest to this manuscript.
Financial Support and Sponsorship
There was no financial support or sponsorship for the study.
References
- Sütterlin S, Bray JE, Miaden MCJ, Tano E. Distribution of class 1 integrons in historic and contemporary collections of human pathogenic Escherichia coli. PLoS One. 2020; 15: e0233315.
- Livermore DM, Walsh TR., Toleman M, Woodford N. Balkan NDM-1: escape or transplant? Lancet of Infectious Disease. 2012; 11:164.
- Ejikeugwu C, Ugwu M, Iriha I, Eze P, Gugu T, Esimone C. Phenotypic detection of metallo-ß-lactamase (MßL) enzyme in Enugu, Southeast Nigeria. American Journal of Biological, Chemical and Pharmaceutical Sciences. 2014; 2 (2): 1-6.
- Aibinu I, Nwanneka T, Odugbemi T. Occurrence of ESBL and MBL in clinical isolates of Pseudomonas aeruginosa from Lagos, Nigeria. Journal of American Science. 2007; 3(4); 81-85.
- Etang UE, Akpan SS, Inyang UC, Akpan NG, Tatfeng MY, Moses AE. Phenotypic detection of multi-drug resistant MBL-producing Gram-negative bacteria isolated from clinical samples of patients attending hospitals in Akwa Ibom State, Nigeria. World Journal of Applied science and Technology. 2022; 14(1b): 104-118.
- Zhao WH, Hu ZQ. Epidemiology and genetics of VIM-type metallo-ß-lactamases in Gram-negative bacilli. Future Microbiology. 2011; 6(3): 317–333.
- Zubair KO, Iregbu KC. Resistance Pattern and Detection of Metallo beta lactamase Genes in Clinical Isolates of Pseudomonas aeruginosa in a Central Nigeria Tertiary Hospital. Niger Journal of Clinical Practice. 2018; 21, 176-182.
- Toleman MA, Simm AM, Murphy TA. Molecular characterization of SPM-1, a novel metallo-ß-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. Journal of Antimicrobial Agents and Chemotherapy. 2002; 50(5): 673–679.
- Sedighi M, Salehi-Abargouei A, Oryan G, Faghri J. Epidemiology of VIM-1-imipenem resistant Pseudomonas aeruginosa in Iran: A systematic review and meta-analysis. Journal of Research in Medical Science. 2014; 19 (9): 899-903.
- Crowder MW, Spencer J, Vila AJ. Metallo-ß-lactmases: novel weaponry for antibiotic resistance in bacteria. Account of Chemical Research. 2006; 39:721–728.
- Sharma M, Pathak S, Srivastava P. Prevalence and antibiogram of extended-spectrum beta-lactamase producing Gram-negative bacteria and further molecular characterization of ß-lactamase-producing Escherichia coli and Klebsiella spp. Journal of Clinical and Diagnostic Research. 2013; 7 (10): 2173-2177.
- Sepehriseresht S, Pourgholi L, Habibi E, Sotoudeh MA, Sattarzadeh T. Detection of VIM and IMP type metallo ß-lactamases in Pseudomonas aeruginosa clinical isolates. Arch Iran Med. 2012; 15(11): 670 -673.
- Adam MA, Elhag WI. Prevalence of metallo-ß-lactamase acquired genes among carbapenems susceptible and resistant Gram-negative clinical isolates using multiplex PCR, Khartoum hospitals, Khartoum Sudan. BMC Infectious Disease. 2018; 18:668.
- Alade TO, Ewaoche SI, Lawani-Luwaji E, Oladapo O. Molecular detection of NDM and VIM genes in Pseudomonas aeruginosa isolates from clinical samples in Bayelsa State, Nigeria. Current Research in Bacteriology. 2022; 15: 8-14.
- Davasena UR., Meenambiga SS. Molecular characterization of antibiotic resistant genes among Gram-negative clinical isolates. Trends in Science. 2022; 19(4): 2687-2698.
- Rossolini GM, Luzzaro F, Migliavacca R. First countrywide survey of acquired metallo-ß-lactamases in gram-negative pathogens in Italy. Journal of Antimicrobial Agents and Chemotherapy. 2008; 52(11): 4023–4029.
- Martinez E, Marquez C, Ingold A, et al. Diverse mobilized class 1 integrons are common in the chromosomes of pathogenic Pseudomonas aeruginosa clinical isolates. Antimicrobial Agents and Chemotherapy. 2012; 56: 2169–2172.
- Clinical and Laboratory Standard Institute. Performance Standard for Antimicrobial Susceptibility Testing. Thirty-third Informational Supplement, CLSI Document M100-S20, Wayne, PA: Clinical and Laboratory Standard Institute; 2023.
- Yong D, Toleman MA, Bell J. Genetic and biochemical characterization of an acquired subgroup B3 metallo-ß-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Journal of Antimicrobial Agents and Chemotherapy. 2012; 56(12): 6154–6159.
- Bandekar N, Vinodkumar CS, Basavarajappa KJ, Prabhakar PJ, Nagaraj P. ß-lactamases mediated resistance amongst Gram negative bacilli in Burn infection. Int J Biol Med Res. 2011; 2(3): 766-770.
- Mohammadzadeh M, Tavakoli M, Mohebi A, Aghayi S. Phenotypic and Genotypic Detection of Metallo-Beta-Lactamases among Imipenem Resistant Gram Negative Isolates J Med Bacteriol. 2016; 5(1-2): 36-42.
- Rameshkumar G, Dhandapani R, Lalitha P, et al. Prevalence and Molecular Characterization of Metallo-beta-Lactamase Producing Gram-Negative Pathogens Causing Eye Infections. Frontiers in Public Health. 2022; 10:Article870354.
- Brooks GF, Carrol KC, Butel JS, Morse SA, Mietzner TA. Jawetz, Melnick and Adelberg’s Medical Microbiology. 26th edition. McGraw Hill Companies Incorporated, USA. 2013; 378-800.
- Deng Y, Bao X, Chen L, et al. Resistance integrons: class 1, 2 and 3 integrons. Annals of Clinical Microbiology and Antimicrobials. 2015; 14(45): 1-11.
- Abdelaziz SM, Aboshanab KM, Yahia IS, Yassien MA, Hassouna N. Correlation between the Antibiotic Resistance Genes and Susceptibility to Antibiotics among the Carbapenem-Resistant Gram-Negative Pathogens. Antibiotics. 2021; 10, 255.
- Siwakoti S, Subedi A, Sharma A, Baral R, Bhattarai NR, Khanal B. Incidence and Outcomes of Multidrug-Resistant Gram-negative Bacteria Infections in Intensive Care Unit from Nepal- a Prospective Cohort Study. Antimicrobial Resistance and Infection Control. 2018; 7, 114.
- Akereuke UE, Onwuezobe IA, Ekuma AE, Edem EN, Etang UE. Molecular profile of metallo-ß-lactamase producing bacterial isolates from clinical samples in University of Uyo Teaching Hospital (UUTH), Uyo, South South, Nigeria. Journal of Research in Clinical Medicine. 2023; (In Press).
- Abdu BA, Egbagba J, Alade T. Antimicrobial Susceptibility Pattern and Bacterial Isolates Profile in Septicaemia Suspected Patients Attending FMC. Yenagoa. International Journal of Research and Scientific Innovation. 2020; 7(4):134-143.
- Ahammad ZS, Sreekrishnan TR., Hands CL, Knapp CW, Graham DW. Increased waterborne blaNDM-1 resistance gene abundances associated with seasonal human pilgrimages to the Upper Ganges River. Environmental Science and Technology. 2014; 48:3014 –3020.
- Ramazanzadeh R., Rouhi S, Shakib P. Molecular detection of extended-spectrum beta-lactamase in isolated bacteria from blood cultures. Journal of Medical Bacteriology. 2015; 4 (1-2): 27-34.
- Ugwu MC, Shariff M, Nnajide CM. et al. Phenotypic and Molecular Characterization of ß-Lactamases among Enterobacterial Uropathogens in Southeastern Nigeria. Canadian Journal of Infectious Disease and Medical Microbiology. 2020; 1-9.
- Isichei-Ukah BO, Enabulele OI. Analysis of class 1 integrons and antibiotic resistance genes in Pseudomonas aeruginosa strains from Benin City. Journal of Applied Science in Environmental Management. 2020; 24(4): 633-637.
- Abdullahi SA, Arzai AH, Yusuf I, et al. Molecular detection of New Delhi metallo beta lactamase 1 (NDM-1) producing bacterial isolates in Kano- Northwestern Nigeria. Annual Research and Review in Biology. 2017; 14(4): 1-6.
- Tewari R, Ganaie F, Venugopal N, Mitra S, Shome R, Shome BR. Occurrence and characterization of genetic determinants of Beta-lactam resistance in Escherichia coli isolates. Infection, Genetics and Evolution. 2022; 100(10): 52-57.
- Mano Y, Saga T, Ishii Y, et al. Molecular analysis of the integrons of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates collected by nationwide surveillance programs across Japan. BMC Microbiology. 2015; 15(41): 1-8.
- Fatemeh F, Maryam N, Ali H, et al. Prevalence of blaNDM, blaPER, blaVEB, blaIMP and blaVIM genes among Acinetobacter baumannii isolated from two hospitals of Tehran, Iran. Scientifica. 2014; 1-6.
- Lauretti L, Riccio ML, Mazzariol A, et al. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Journal of Antimicrobial Agents and Chemotherapy. 1999; 43:1584-90.
- Karlowsky JA, Kazmierczak KM, de Jonge BLM, Hackel MA, Sahm DF, Bradford PA. In vitro activity of aztreonam-avibactam against enterobacteriaceae and Pseudomonas aeruginosa isolated by clinical laboratories in 40 Countries from 2012 to 2015. Journal of Antimicrobial Chemotherapy. 2017; 61:9-17.
- Umo AN, Etang UE, Ama VO, Moses AE. Phenotypic detection of multi-drug resistant extended spectrum beta-lactamase-producing Gram-negative clinical bacteria in health care facilities in Akwa Ibom State, Nigeria. World Journal of Applied Science and Technology. 2021; 13(1): 114-123.
- Onwuezobe IA, Akereuke UE, Ekuma EA. Occurrence and associated risk factors of metallo-beta-lactamase producing agents of infection among patients in Uyo, Southern Nigeria. Microbiology and Infectious Diseases. 2023; 7(1): 1-6.
- Uyanga FZ, Ekundayo EO, Nwankwo EO. bla TEM, bla SHV and bla CTX-M-15 Extended Spectrum Beta-lactamase Produced by Acinetobacter baumanii, Enterobacter clocae and Proteus mirabilis from Pregnant Women in Three Secondary Health Care Facilities in South-south, Nigeria. Journal of Advances in Microbiology. 2019; 18(1): 1-9.